|
chr9_+_136673534
|
9.509
|
|
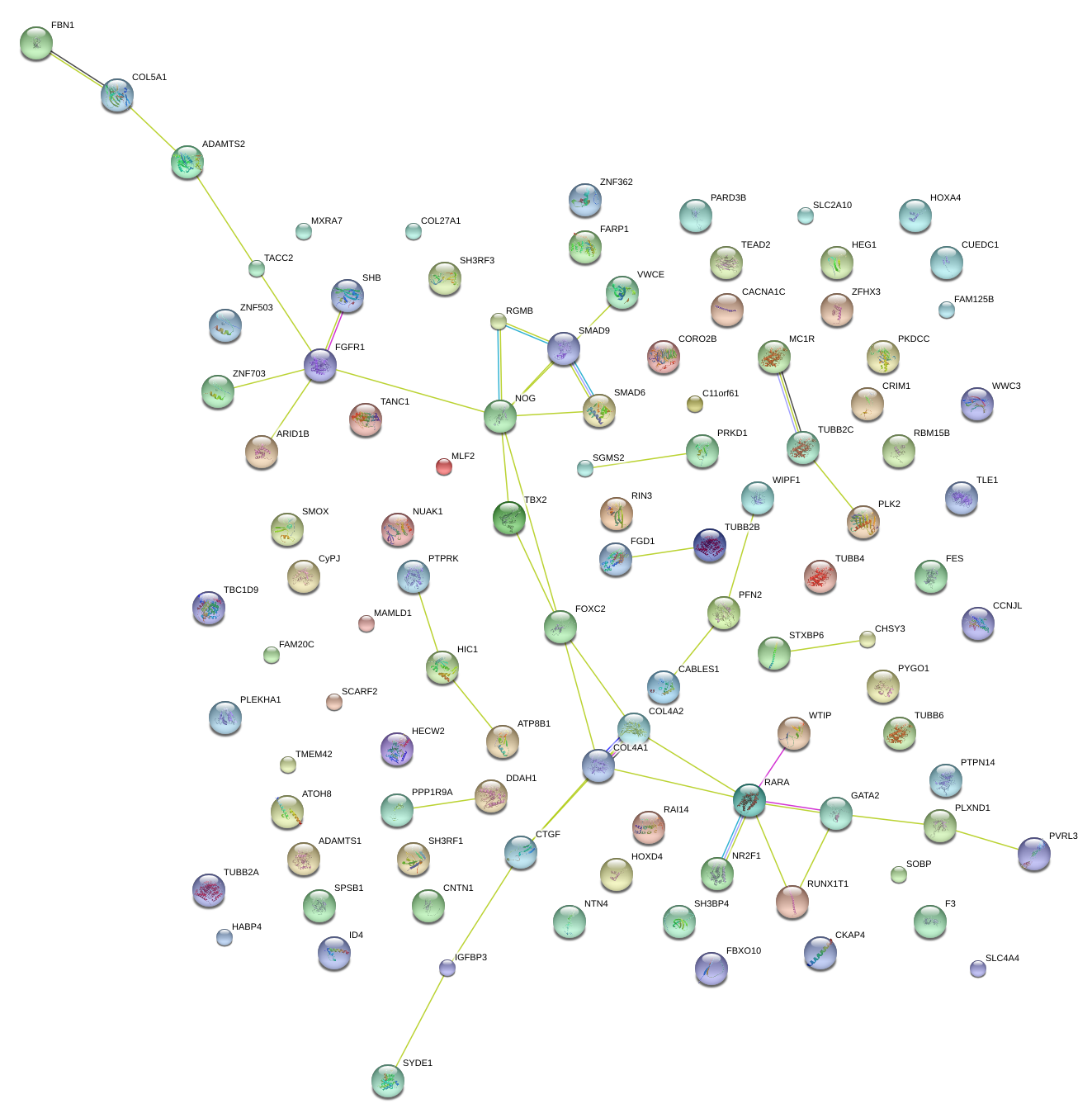
COL5A1
|
collagen, type V, alpha 1
|
|
chr9_+_136673464
|
7.949
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr15_-_46724360
|
7.556
|
|
FBN1
|
fibrillin 1
|
|
chr17_+_56832255
|
7.085
|
|
TBX2
|
T-box 2
|
|
chr17_+_56831951
|
6.969
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr3_-_126257425
|
6.423
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr15_+_64782468
|
6.277
|
|
SMAD6
|
SMAD family member 6
|
|
chr14_+_92049396
|
5.877
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr2_+_85834111
|
5.808
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr2_+_36436873
|
5.576
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr6_+_107917965
|
5.454
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr20_+_44771532
|
5.450
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr2_+_159533329
|
5.443
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr9_-_83493415
|
5.288
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr14_-_24588916
|
5.207
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr3_-_130807865
|
5.169
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr19_+_39664706
|
5.167
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr5_+_92946348
|
5.088
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr15_-_53667800
|
4.989
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr19_+_15079141
|
4.972
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr18_-_53621277
|
4.934
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr3_+_51403760
|
4.872
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr10_-_76829857
|
4.832
|
|
ZNF503
|
zinc finger protein 503
|
|
chr8_+_37672427
|
4.737
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr8_-_38444982
|
4.714
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr22_-_19122107
|
4.472
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr2_+_205118710
|
4.446
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr4_+_72271218
|
4.427
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_+_56832492
|
4.338
|
|
TBX2
|
T-box 2
|
|
chr1_-_94779727
|
4.216
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr12_-_105165805
|
4.195
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr17_-_72218661
|
4.174
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr2_+_36436316
|
4.134
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr14_-_29466562
|
4.118
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr11_-_124175467
|
4.041
|
|
C11orf61
|
chromosome 11 open reading frame 61
|
|
chr2_+_235525355
|
4.021
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr3_+_44878380
|
3.976
|
NM_144638
|
TMEM42
|
transmembrane protein 42
|
|
chr1_+_33494745
|
3.967
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr12_+_2032649
|
3.960
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr6_+_1336077
|
3.959
|
|
|
|
|
chr11_-_60819110
|
3.946
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr7_+_288051
|
3.920
|
NM_020223
|
FAM20C
|
family with sequence similarity 20, member C
|
|
chr15_+_89228668
|
3.910
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr1_-_85703198
|
3.905
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr16_-_71639670
|
3.876
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr16_+_85158357
|
3.819
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr5_+_98132898
|
3.801
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chrX_+_149282097
|
3.789
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr17_+_35751796
|
3.749
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr2_+_42128521
|
3.748
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr16_-_71650034
|
3.737
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr9_-_37566248
|
3.721
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr11_-_124175493
|
3.711
|
NM_024631
|
C11orf61
|
chromosome 11 open reading frame 61
|
|
chr12_-_105056577
|
3.682
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chrX_-_54538591
|
3.649
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr11_-_60819273
|
3.624
|
NM_152718
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr6_+_19945578
|
3.616
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr2_+_205118961
|
3.609
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr14_+_54102518
|
3.580
|
|
LOC644925
|
hypothetical LOC644925
|
|
chr2_+_205118801
|
3.575
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr5_+_129268352
|
3.498
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr3_+_112273352
|
3.493
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr4_+_108965167
|
3.463
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_151171190
|
3.458
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr6_-_132314004
|
3.455
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr20_+_4077466
|
3.454
|
|
SMOX
|
spermine oxidase
|
|
chr3_+_112273464
|
3.442
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr8_-_38444675
|
3.412
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr2_+_109112428
|
3.394
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chrX_+_9943793
|
3.380
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chr7_+_94374862
|
3.372
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr17_+_1906262
|
3.361
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr7_-_27136876
|
3.360
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr9_+_98252200
|
3.321
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr9_+_128128913
|
3.320
|
NM_001011703
NM_033446
|
FAM125B
|
family with sequence similarity 125, member B
|
|
chr2_-_197165495
|
3.316
|
NM_020760
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
|
|
chr19_-_54557450
|
3.297
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr10_+_123862430
|
3.225
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr5_-_178704934
|
3.215
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr1_-_212791467
|
3.206
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr21_-_27139143
|
3.193
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr5_-_159672060
|
3.180
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr13_+_109757593
|
3.175
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr4_-_141896920
|
3.144
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr18_+_18969724
|
3.124
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr6_-_128883125
|
3.120
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr9_+_115958051
|
3.098
|
NM_032888
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr10_+_124124196
|
3.093
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr8_-_93184629
|
3.074
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_-_53387493
|
3.070
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr7_-_5427367
|
3.050
|
|
|
|
|
chr5_+_34692124
|
3.042
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr16_+_88517187
|
3.042
|
NM_006086
|
TUBB3
|
tubulin, beta 3
|
|
chr6_+_157141565
|
3.019
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr1_+_9275542
|
3.019
|
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr17_+_52026058
|
3.006
|
NM_005450
|
NOG
|
noggin
|
|
chr2_-_175255717
|
2.996
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr17_-_72218535
|
2.979
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr13_-_109757442
|
2.973
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr3_-_129689394
|
2.972
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr7_-_45927263
|
2.971
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr7_+_288195
|
2.956
|
|
FAM20C
|
family with sequence similarity 20, member C
|
|
chr15_+_66658626
|
2.952
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr13_+_97593711
|
2.947
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr5_-_57791544
|
2.938
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr3_-_151171576
|
2.935
|
|
PFN2
|
profilin 2
|
|
chr13_-_36392370
|
2.922
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr9_-_38059082
|
2.922
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr12_-_94708477
|
2.915
|
|
NTN4
|
netrin 4
|
|
chr8_-_48813256
|
2.913
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr20_+_11819364
|
2.884
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr5_+_34692352
|
2.883
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr5_-_178705036
|
2.849
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr5_+_34692239
|
2.839
|
|
RAI14
|
retinoic acid induced 14
|
|
chr10_-_76831545
|
2.839
|
|
ZNF503
|
zinc finger protein 503
|
|
chr10_-_76831430
|
2.838
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr1_-_95165037
|
2.832
|
|
CNN3
|
calponin 3, acidic
|
|
chr10_-_725522
|
2.826
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr13_+_30378311
|
2.823
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr8_+_30361485
|
2.821
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr3_+_171558188
|
2.819
|
|
SKIL
|
SKI-like oncogene
|
|
chr1_+_9275518
|
2.813
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr10_-_118491971
|
2.812
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr8_-_48813236
|
2.797
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr12_-_94708641
|
2.793
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr3_+_160964574
|
2.793
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr22_-_19122047
|
2.789
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr2_+_241586927
|
2.785
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr10_-_76831142
|
2.779
|
|
ZNF503
|
zinc finger protein 503
|
|
chr8_-_38444398
|
2.743
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr2_+_42129440
|
2.724
|
|
|
|
|
chr2_-_45090025
|
2.712
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr3_+_189354167
|
2.686
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr4_-_22126437
|
2.668
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr12_-_15833617
|
2.666
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr2_+_241023908
|
2.666
|
|
GPC1
|
glypican 1
|
|
chr19_-_15172773
|
2.633
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr6_+_151603346
|
2.627
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr12_+_79995927
|
2.621
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr10_+_116571485
|
2.619
|
NM_001135051
NM_020940
|
FAM160B1
|
family with sequence similarity 160, member B1
|
|
chr22_-_32646325
|
2.606
|
NM_004737
NM_133642
|
LARGE
|
like-glycosyltransferase
|
|
chr7_-_122313646
|
2.594
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr9_-_135847106
|
2.585
|
NM_001134398
NM_003371
|
VAV2
|
vav 2 guanine nucleotide exchange factor
|
|
chr5_+_60663842
|
2.577
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr19_+_1200882
|
2.573
|
|
MIDN
|
midnolin
|
|
chr5_+_72957736
|
2.568
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr8_+_37773991
|
2.565
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr21_-_27139563
|
2.559
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr15_+_30797466
|
2.549
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr18_+_2896956
|
2.512
|
|
EMILIN2
|
elastin microfibril interfacer 2
|
|
chr17_-_1306160
|
2.512
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr3_-_121652183
|
2.494
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr8_-_18915443
|
2.471
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr2_-_127580975
|
2.468
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr1_-_32940943
|
2.462
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr7_-_139987044
|
2.437
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr10_+_104394185
|
2.434
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr20_-_4177519
|
2.433
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr1_+_199884152
|
2.417
|
|
NAV1
|
neuron navigator 1
|
|
chr17_+_17525571
|
2.416
|
|
|
|
|
chr11_-_65841077
|
2.402
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr14_+_51188289
|
2.398
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr16_-_69277194
|
2.396
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr4_+_151218862
|
2.396
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr10_-_105410840
|
2.390
|
|
|
|
|
chr15_-_53822468
|
2.386
|
NM_173814
|
PRTG
|
protogenin
|
|
chr6_-_117193505
|
2.380
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr5_-_121441866
|
2.378
|
|
LOX
|
lysyl oxidase
|
|
chr19_+_16296632
|
2.373
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr10_+_59942874
|
2.362
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr14_-_102593384
|
2.361
|
NM_006035
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like)
|
|
chr11_-_65396775
|
2.358
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr10_+_128583984
|
2.347
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr17_-_19711580
|
2.345
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr10_+_122206455
|
2.341
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr16_-_69277355
|
2.339
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr15_+_65145215
|
2.333
|
|
SMAD3
|
SMAD family member 3
|
|
chr2_-_144991386
|
2.331
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr20_+_8060762
|
2.320
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr4_-_158111995
|
2.314
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr12_+_2032964
|
2.312
|
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr6_+_3696224
|
2.306
|
|
|
|
|
chr10_-_100017933
|
2.298
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr22_-_34754343
|
2.292
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr10_+_94439658
|
2.289
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr14_+_99329198
|
2.287
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr15_-_58671929
|
2.282
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_-_88084387
|
2.275
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr2_+_236068029
|
2.255
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr11_-_1549719
|
2.255
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr19_+_1199548
|
2.254
|
NM_177401
|
MIDN
|
midnolin
|
|
chr9_-_25668230
|
2.244
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr14_+_68796621
|
2.238
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr15_+_72006015
|
2.238
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr9_-_38059207
|
2.235
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr13_+_26029799
|
2.234
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr1_+_179148935
|
2.230
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr6_+_1335067
|
2.228
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr2_+_54536780
|
2.222
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr5_-_107744971
|
2.213
|
|
FBXL17
|
F-box and leucine-rich repeat protein 17
|